|
chr2_-_217560247
|
16.584
|
NM_000599
|
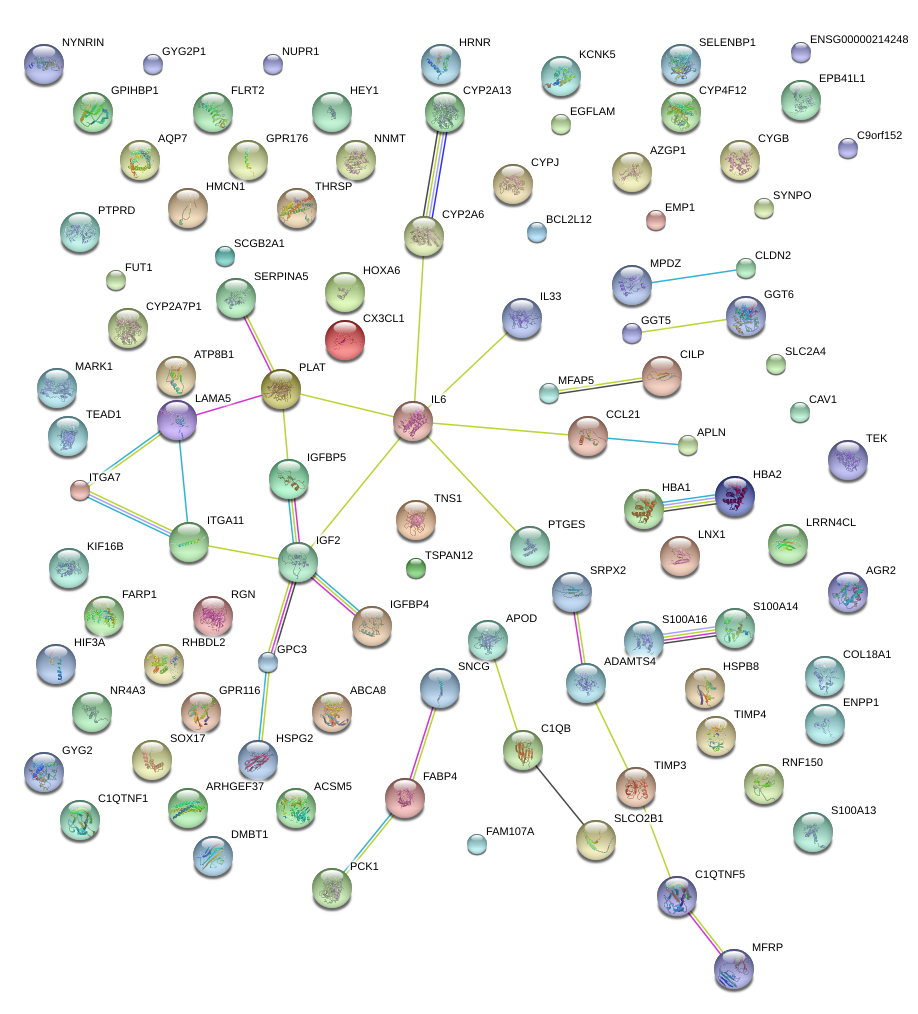
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr7_-_16844606
|
9.908
|
NM_006408
|
AGR2
|
anterior gradient 2 homolog (Xenopus laevis)
|
|
chrX_-_133119475
|
9.707
|
|
GPC3
|
glypican 3
|
|
chr8_-_82395401
|
9.345
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr12_-_56106061
|
8.827
|
NM_001144997
|
ITGA7
|
integrin, alpha 7
|
|
chr15_-_65503786
|
8.812
|
NM_003613
|
CILP
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase
|
|
chr17_+_77030471
|
8.442
|
NM_198593
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr1_-_153588789
|
8.284
|
NM_020672
|
S100A14
|
S100 calcium binding protein A14
|
|
chrX_+_99899162
|
8.271
|
NM_014467
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr20_+_56136136
|
8.179
|
NM_002591
|
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble)
|
|
chr8_+_144295067
|
8.097
|
NM_178172
|
GPIHBP1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1
|
|
chr1_-_151345156
|
8.066
|
NM_003944
|
SELENBP1
|
selenium binding protein 1
|
|
chrX_-_128788913
|
7.795
|
NM_017413
|
APLN
|
apelin
|
|
chrX_-_133119624
|
7.662
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr14_+_85996454
|
7.622
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr7_+_116165043
|
7.615
|
NM_001172895
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr11_+_77774902
|
7.544
|
NM_003251
|
THRSP
|
thyroid hormone responsive
|
|
chr8_-_42065037
|
7.497
|
|
PLAT
|
plasminogen activator, tissue
|
|
chr7_+_116164838
|
7.256
|
NM_001753
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr5_+_38445577
|
7.209
|
NM_182801
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr6_-_46922533
|
7.098
|
NM_015234
|
GPR116
|
G protein-coupled receptor 116
|
|
chr19_+_46801632
|
7.076
|
NM_022462
NM_152796
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|
|
chrX_+_2746810
|
7.052
|
NM_001079855
NM_001184702
NM_001184703
NM_001184704
NM_003918
|
GYG2
|
glycogenin 2
|
|
chr7_+_116165089
|
7.033
|
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr16_+_57406398
|
6.985
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr11_-_62457001
|
6.844
|
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr17_-_66951381
|
6.762
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr16_+_222845
|
6.626
|
NM_000517
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr17_+_38599651
|
6.452
|
NM_001552
|
IGFBP4
|
insulin-like growth factor binding protein 4
|
|
chr16_+_226678
|
6.369
|
NM_000558
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr17_-_74533623
|
6.253
|
NM_134268
|
CYGB
|
cytoglobin
|
|
chr17_-_4463816
|
6.221
|
NM_001122890
NM_153338
|
GGT6
|
gamma-glutamyltransferase 6
|
|
chr8_-_80679897
|
6.187
|
NM_001040708
NM_012258
|
HEY1
|
hairy/enhancer-of-split related with YRPW motif 1
|
|
chr10_+_88718287
|
6.176
|
NM_003087
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1)
|
|
chr1_-_153585504
|
6.091
|
NM_080388
|
S100A16
|
S100 calcium binding protein A16
|
|
chr11_+_74862031
|
6.031
|
NM_001145212
NM_007256
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1
|
|
chr12_+_13349716
|
5.883
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr15_-_40213087
|
5.830
|
NM_007223
|
GPR176
|
G protein-coupled receptor 176
|
|
chr7_-_27187367
|
5.801
|
NM_024014
|
HOXA6
|
homeobox A6
|
|
chr14_+_85996512
|
5.768
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr7_+_22766765
|
5.734
|
NM_000600
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr9_+_27109138
|
5.729
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr11_-_119211526
|
5.693
|
|
C1QTNF5
MFRP
|
C1q and tumor necrosis factor related protein 5
membrane frizzled-related protein
|
|
chr19_-_49258646
|
5.523
|
NM_000148
|
FUT1
|
fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group)
|
|
chr12_+_13349601
|
5.519
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr19_+_46806855
|
5.499
|
NM_152794
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|
|
chr14_+_24867926
|
5.429
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr22_-_24640850
|
5.407
|
NM_001099781
NM_001099782
NM_004121
|
GGT5
|
gamma-glutamyltransferase 5
|
|
chr8_-_42065193
|
5.309
|
NM_000930
NM_033011
|
PLAT
|
plasminogen activator, tissue
|
|
chr9_+_6215785
|
5.290
|
NM_001199640
NM_001199641
NM_033439
|
IL33
|
interleukin 33
|
|
chr1_+_185703470
|
5.196
|
NM_031935
|
HMCN1
|
hemicentin 1
|
|
chr14_+_95047792
|
5.168
|
|
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5
|
|
chr11_+_12960793
|
5.156
|
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr19_+_15783827
|
5.150
|
NM_023944
|
CYP4F12
|
cytochrome P450, family 4, subfamily F, polypeptide 12
|
|
chr12_+_13349720
|
5.124
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr9_-_13279562
|
5.093
|
|
MPDZ
|
multiple PDZ domain protein
|
|
chrX_+_46937744
|
5.087
|
NM_004683
NM_152869
|
RGN
|
regucalcin (senescence marker protein-30)
|
|
chr3_-_58563070
|
5.080
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr20_+_34742656
|
4.947
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chrX_+_46937807
|
4.872
|
|
RGN
|
regucalcin (senescence marker protein-30)
|
|
chr3_-_12200339
|
4.846
|
|
TIMP4
|
TIMP metallopeptidase inhibitor 4
|
|
chr16_-_28550324
|
4.670
|
NM_001042483
NM_012385
|
NUPR1
|
nuclear protein, transcriptional regulator, 1
|
|
chr17_+_7184978
|
4.618
|
NM_001042
|
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4
|
|
chrX_+_46937889
|
4.576
|
|
RGN
|
regucalcin (senescence marker protein-30)
|
|
chr2_-_218843541
|
4.562
|
|
TNS1
|
tensin 1
|
|
chr3_-_12200850
|
4.533
|
NM_003256
|
TIMP4
|
TIMP metallopeptidase inhibitor 4
|
|
chr1_-_39407449
|
4.506
|
NM_017821
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila)
|
|
chr11_-_62457370
|
4.484
|
NM_203422
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr9_-_8733893
|
4.464
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr12_+_119616594
|
4.428
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr7_-_99573637
|
4.409
|
NM_001185
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding
|
|
chr18_-_55470279
|
4.347
|
NM_005603
|
ATP8B1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1
|
|
chr11_-_2159472
|
4.316
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr12_-_8815370
|
4.262
|
NM_003480
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr5_+_148961035
|
4.236
|
NM_001001669
|
ARHGEF37
|
Rho guanine nucleotide exchange factor (GEF) 37
|
|
chr11_+_114166534
|
4.152
|
NM_006169
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr9_-_33402505
|
4.130
|
NM_001170
|
AQP7
|
aquaporin 7
|
|
chr3_-_195306274
|
4.113
|
|
APOD
|
apolipoprotein D
|
|
chr19_-_41356193
|
4.105
|
NM_000762
|
CYP2A6
|
cytochrome P450, family 2, subfamily A, polypeptide 6
|
|
chr22_+_33196801
|
4.093
|
NM_000362
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr8_+_55370494
|
4.044
|
NM_022454
|
SOX17
|
SRY (sex determining region Y)-box 17
|
|
chr6_-_39197225
|
4.014
|
NM_003740
|
KCNK5
|
potassium channel, subfamily K, member 5
|
|
chr11_+_61976139
|
4.008
|
NM_002407
|
SCGB2A1
|
secretoglobin, family 2A, member 1
|
|
chr9_-_112970412
|
3.999
|
NM_001012993
|
C9orf152
|
chromosome 9 open reading frame 152
|
|
chr1_-_22263676
|
3.956
|
NM_005529
|
HSPG2
|
heparan sulfate proteoglycan 2
|
|
chr9_+_102584136
|
3.913
|
NM_006981
NM_173199
NM_173200
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr7_-_120498128
|
3.865
|
NM_012338
|
TSPAN12
|
tetraspanin 12
|
|
chr1_-_153599969
|
3.804
|
NM_001024211
|
S100A13
|
S100 calcium binding protein A13
|
|
chr1_-_161168816
|
3.711
|
NM_005099
|
ADAMTS4
|
ADAM metallopeptidase with thrombospondin type 1 motif, 4
|
|
chr16_+_20420854
|
3.677
|
NM_017888
|
ACSM5
|
acyl-CoA synthetase medium-chain family member 5
|
|
chr21_+_46875402
|
3.633
|
NM_030582
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr1_+_220701547
|
3.633
|
NM_018650
|
MARK1
|
MAP/microtubule affinity-regulating kinase 1
|
|
chr1_+_43735664
|
3.633
|
NM_144626
|
TMEM125
|
transmembrane protein 125
|
|
chr1_+_22979681
|
3.616
|
NM_000491
|
C1QB
|
complement component 1, q subcomponent, B chain
|
|
chr6_+_132129154
|
3.603
|
NM_006208
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr4_-_142054431
|
3.593
|
NM_020724
|
RNF150
|
ring finger protein 150
|
|
chr9_-_132515290
|
3.583
|
NM_004878
|
PTGES
|
prostaglandin E synthase
|
|
chr10_+_124320180
|
3.581
|
NM_004406
NM_007329
NM_017579
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chrX_+_106171328
|
3.545
|
|
CLDN2
|
claudin 2
|
|
chr13_+_98795410
|
3.523
|
NM_001001715
NM_005766
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr1_-_153600714
|
3.497
|
NM_005979
|
S100A13
|
S100 calcium binding protein A13
|
|
chr20_-_60942300
|
3.476
|
NM_005560
|
LAMA5
|
laminin, alpha 5
|
|
chr5_+_150020201
|
3.470
|
NM_001109974
NM_007286
|
SYNPO
|
synaptopodin
|
|
chr15_-_68724434
|
3.470
|
NM_001004439
|
ITGA11
|
integrin, alpha 11
|
|
chr9_-_34710062
|
3.465
|
NM_002989
|
CCL21
|
chemokine (C-C motif) ligand 21
|
|
chr17_-_15164043
|
3.437
|
NM_153322
|
PMP22
|
peripheral myelin protein 22
|
|
chr1_-_16344438
|
3.427
|
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr20_-_6034617
|
3.389
|
NM_152611
|
LRRN4
|
leucine rich repeat neuronal 4
|
|
chr13_+_98795468
|
3.382
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr17_-_46671043
|
3.376
|
NM_002147
|
HOXB5
|
homeobox B5
|
|
chr19_-_33716609
|
3.372
|
NM_019849
|
SLC7A10
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10
|
|
chr2_-_216300444
|
3.362
|
|
FN1
|
fibronectin 1
|
|
chrX_+_99899388
|
3.344
|
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr16_-_86542463
|
3.321
|
|
LOC400550
|
uncharacterized LOC400550
|
|
chr17_-_76975932
|
3.306
|
NM_005567
|
LGALS3BP
|
lectin, galactoside-binding, soluble, 3 binding protein
|
|
chr20_+_42194736
|
3.305
|
NM_016276
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr19_+_36157714
|
3.283
|
NM_007000
|
UPK1A
|
uroplakin 1A
|
|
chr16_-_52581713
|
3.195
|
NM_001146188
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr10_+_82173574
|
3.176
|
NM_001243778
NM_001243782
|
C10orf58
|
chromosome 10 open reading frame 58
|
|
chr7_+_22767101
|
3.167
|
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr14_-_76447323
|
3.147
|
|
TGFB3
|
transforming growth factor, beta 3
|
|
chr1_-_160854869
|
3.142
|
NM_017625
|
ITLN1
|
intelectin 1 (galactofuranose binding)
|
|
chr9_-_130616957
|
3.082
|
|
ENG
|
endoglin
|
|
chrX_+_106161589
|
3.076
|
NM_001171095
|
CLDN2
|
claudin 2
|
|
chr3_-_50340979
|
3.074
|
NM_007312
NM_033159
NM_153282
NM_153283
NM_153285
|
HYAL1
|
hyaluronoglucosaminidase 1
|
|
chr9_+_136399877
|
3.047
|
NM_014694
|
ADAMTSL2
|
ADAMTS-like 2
|
|
chr14_-_76447445
|
3.024
|
|
TGFB3
|
transforming growth factor, beta 3
|
|
chr19_-_39523132
|
3.014
|
NM_178820
|
FBXO27
|
F-box protein 27
|
|
chr5_+_176516550
|
3.011
|
NM_022963
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr18_-_5892102
|
3.004
|
NM_001080209
|
TMEM200C
|
transmembrane protein 200C
|
|
chr11_-_6341708
|
2.962
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr16_-_19896105
|
2.951
|
NM_016235
|
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B
|
|
chr11_+_74870824
|
2.919
|
NM_001145211
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1
|
|
chr11_-_2016743
|
2.915
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr9_-_130616592
|
2.909
|
|
ENG
|
endoglin
|
|
chr1_+_33231227
|
2.894
|
NM_001198972
NM_001198973
|
KIAA1522
|
KIAA1522
|
|
chr5_-_148758787
|
2.890
|
NM_014443
|
IL17B
|
interleukin 17B
|
|
chr12_-_8815479
|
2.869
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr10_+_135160843
|
2.847
|
NM_001145201
NM_145202
|
PRAP1
|
proline-rich acidic protein 1
|
|
chr14_+_95047758
|
2.837
|
|
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5
|
|
chr3_-_149421059
|
2.827
|
NM_001168278
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chrX_+_134124967
|
2.827
|
NM_001163438
|
CXorf69
|
chromosome X open reading frame 69
|
|
chr1_+_201979689
|
2.825
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr14_-_76447235
|
2.816
|
|
TGFB3
|
transforming growth factor, beta 3
|
|
chr7_+_102553343
|
2.807
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr21_+_33784751
|
2.804
|
NM_058187
|
C21orf63
|
chromosome 21 open reading frame 63
|
|
chr1_+_162602198
|
2.802
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr2_+_220325533
|
2.799
|
NM_001173476
|
SPEG
|
SPEG complex locus
|
|
chr1_-_23810627
|
2.796
|
|
ASAP3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3
|
|
chr14_-_76448091
|
2.777
|
NM_003239
|
TGFB3
|
transforming growth factor, beta 3
|
|
chr17_-_39661803
|
2.735
|
NM_002274
NM_153490
|
KRT13
|
keratin 13
|
|
chr22_+_30792929
|
2.727
|
NM_001204204
NM_012429
NM_033382
|
SEC14L2
|
SEC14-like 2 (S. cerevisiae)
|
|
chr9_-_33402463
|
2.723
|
|
AQP7
|
aquaporin 7
|
|
chr1_-_153599743
|
2.711
|
NM_001024212
|
S100A13
|
S100 calcium binding protein A13
|
|
chr19_-_35633354
|
2.703
|
|
LGI4
|
leucine-rich repeat LGI family, member 4
|
|
chr12_-_8815266
|
2.701
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr14_+_95047730
|
2.613
|
NM_000624
|
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5
|
|
chr14_+_96858436
|
2.590
|
NM_152327
|
AK7
|
adenylate kinase 7
|
|
chr8_-_22785228
|
2.589
|
NM_144962
|
PEBP4
|
phosphatidylethanolamine-binding protein 4
|
|
chrX_+_152770083
|
2.588
|
|
BGN
|
biglycan
|
|
chr1_+_153600859
|
2.585
|
NM_006271
|
S100A1
|
S100 calcium binding protein A1
|
|
chr9_-_130617009
|
2.575
|
NM_000118
NM_001114753
|
ENG
|
endoglin
|
|
chr20_+_9049660
|
2.573
|
NM_001172646
|
PLCB4
|
phospholipase C, beta 4
|
|
chr1_+_95558072
|
2.522
|
NM_001199679
|
TMEM56
|
transmembrane protein 56
|
|
chr17_+_37894212
|
2.516
|
|
GRB7
|
growth factor receptor-bound protein 7
|
|
chrX_-_13956445
|
2.516
|
|
|
|
|
chr9_-_130616760
|
2.510
|
|
ENG
|
endoglin
|
|
chr20_+_56725982
|
2.491
|
NM_178456
|
C20orf85
|
chromosome 20 open reading frame 85
|
|
chr21_+_47531369
|
2.490
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr12_+_16506350
|
2.476
|
NM_145764
|
MGST1
|
microsomal glutathione S-transferase 1
|
|
chr10_+_5454516
|
2.460
|
NM_001047160
|
NET1
|
neuroepithelial cell transforming 1
|
|
chr1_-_153538305
|
2.455
|
NM_005978
|
S100A2
|
S100 calcium binding protein A2
|
|
chr1_+_203595897
|
2.421
|
NM_001001396
NM_001684
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr1_-_23810721
|
2.414
|
NM_001143778
NM_017707
|
ASAP3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3
|
|
chrX_+_13587693
|
2.411
|
NM_001167890
NM_015507
|
EGFL6
|
EGF-like-domain, multiple 6
|
|
chr4_+_183245136
|
2.410
|
NM_001080477
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr8_-_81787015
|
2.394
|
NM_001033723
|
ZNF704
|
zinc finger protein 704
|
|
chr8_-_12973752
|
2.386
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr12_+_7169192
|
2.382
|
|
|
|
|
chr6_+_31982538
|
2.378
|
NM_001242823
NM_001252204
NM_007293
NM_001002029
|
LOC100293534
C4A
C4B
|
complement C4-B-like
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr3_-_48471401
|
2.375
|
NM_001130082
|
PLXNB1
|
plexin B1
|
|
chr2_-_228582698
|
2.370
|
NM_025243
|
SLC19A3
|
solute carrier family 19, member 3
|
|
chr16_-_28550227
|
2.364
|
|
NUPR1
|
nuclear protein, transcriptional regulator, 1
|
|
chr22_+_40390935
|
2.343
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr14_-_61190458
|
2.332
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr12_+_124773709
|
2.311
|
NM_181709
|
FAM101A
|
family with sequence similarity 101, member A
|
|
chr20_-_44168128
|
2.310
|
NM_080827
|
WFDC6
|
WAP four-disulfide core domain 6
|
|
chr11_-_57089670
|
2.244
|
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa
|
|
chr2_-_233877920
|
2.220
|
NM_019850
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chr10_+_106034886
|
2.211
|
NM_001191014
NM_001191015
|
GSTO2
|
glutathione S-transferase omega 2
|
|
chr17_-_41910439
|
2.209
|
NM_001932
|
MPP3
|
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3)
|
|
chr9_-_95244750
|
2.196
|
NM_001193335
NM_017680
|
ASPN
|
asporin
|
|
chr1_-_161059353
|
2.191
|
NM_030916
|
PVRL4
|
poliovirus receptor-related 4
|
|
chr6_+_132129195
|
2.147
|
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr9_+_74764266
|
2.139
|
NM_001242505
NM_001242506
NM_004293
|
GDA
|
guanine deaminase
|
|
chr8_-_13133956
|
2.136
|
|
|
|
|
chr22_-_31536339
|
2.130
|
NM_015715
|
PLA2G3
|
phospholipase A2, group III
|
|
chr8_-_21988518
|
2.100
|
NM_005144
NM_018411
|
HR
|
hairless homolog (mouse)
|
|
chr1_+_101185195
|
2.099
|
NM_001078
NM_001199834
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr11_+_6260321
|
2.095
|
NM_001037329
|
CNGA4
|
cyclic nucleotide gated channel alpha 4
|